Projects
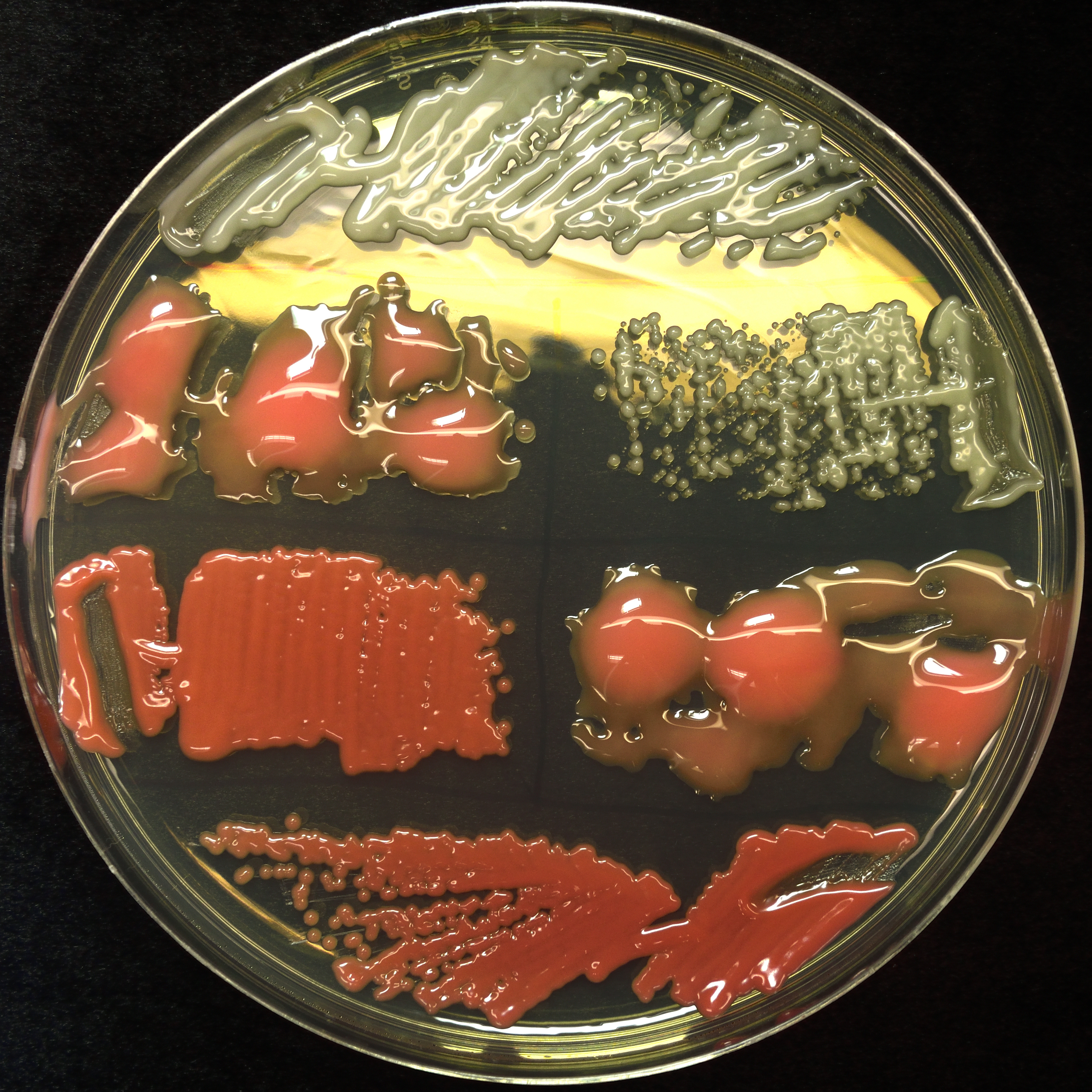
Genetics of phyllosphere commensalism in Aureobasidium pullulans

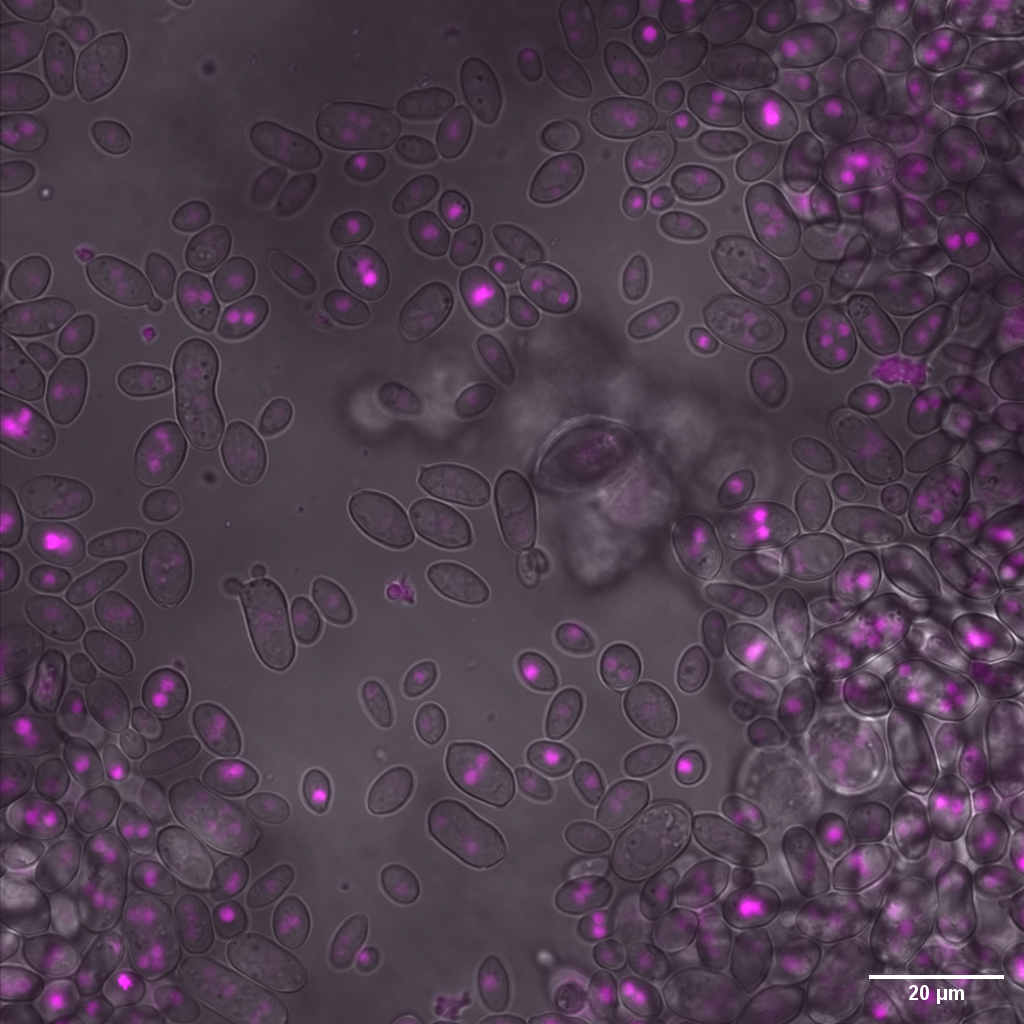
Aureobasidium pullulans is an extremely common member of the plant microbiome, and has characterisitics such as frequent recombination, significant phenotypic variation, ease of isolation and identification, and genetic tractability which makes it an ideal organism to study the ecology and physiology of eukaryotic microbes on healthy plants. I am working with collaborators to develop a genome-sequenced collection of A. pullulans isolates for conducting GWAS and other genetic studies of traits important to plant colonization and other interesting aspects of the yeast, such as nuclear division and budding processes.
Engineering phyllosphere yeasts
Yeasts, of many different genera, are abundant and persistent members of the microbial communities on plant leaves. Genetic manipulation of yeasts may be useful to inform the fundemental processes involved in community assembly and host-microbe interactions, and may be leveraged to improve plant resiliency under stress.
Effects of immune-deficient genotypes of Arabidopsis thaliana on eukaryotic phyllosphere microbiomes
Several mutants of Arabidopsis thaliana which are missing receptors for recognizing pathogens can develop a harmful assembly of microorganisms in their leaves, called dysbiosis. We are interested in the role fungi may play in the process of dysbiosis.
Associations of Linnemannia elongata with Arabidopsis thalina roots
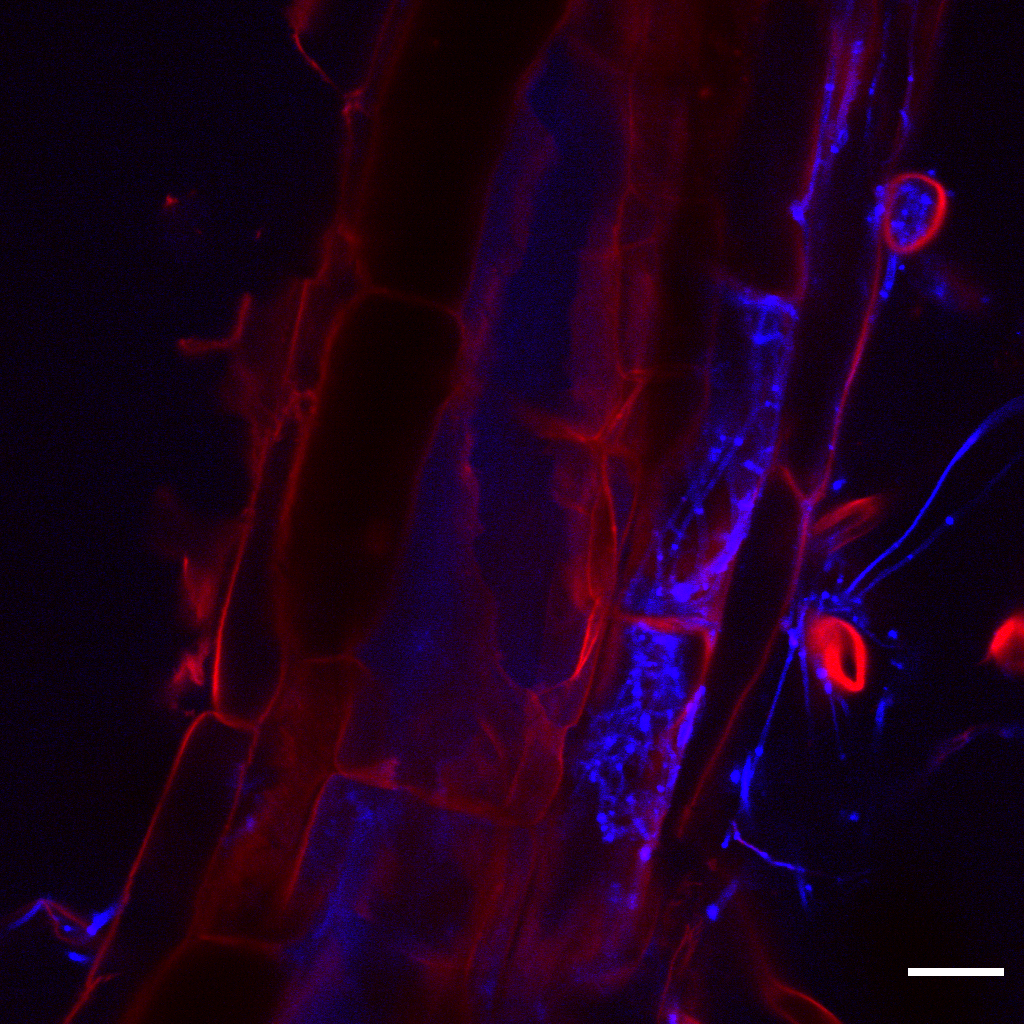
Many fungi and close associations with host plants, and alter their physiology in ways that affect their fitness and agronomic traits. We examined the role of the abundant soil fungus Linnemannia elongata and its associated endohyphal bacteria on the growth of the model plant Arabidopsis thaliana.
Collaborators: Dr. Natalie Vande Pol
Interactions of bacterial and fungal endophytes with plant disease
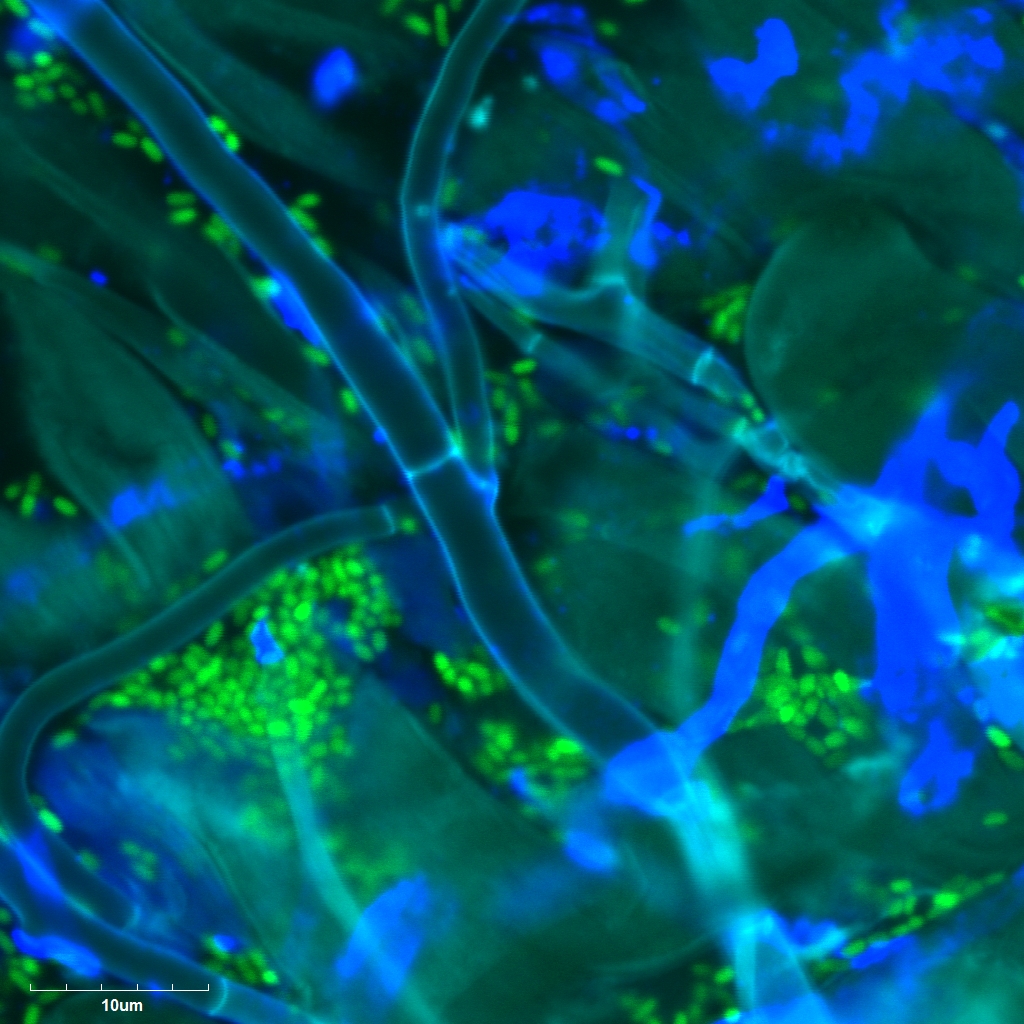
Bacteria and fungi occupy tissues of plants and can affect phenotypes of their host, often in response to stress. How these communities colonize plant roots with respect to other symbionts and plant disease state is not well explained. Using co-inoculation of bacteria and fungi on Brachypodium distachyon, and infection with barley yellow dwarf virus, we have worked to explore the factors influencing microbial community assembly.
Collaborators: Natalie Golematis, Dr. Carolyn Malmstrom, Ellen Cole, Kota Nakasato
Nematode vectoring of fungal endobacteria
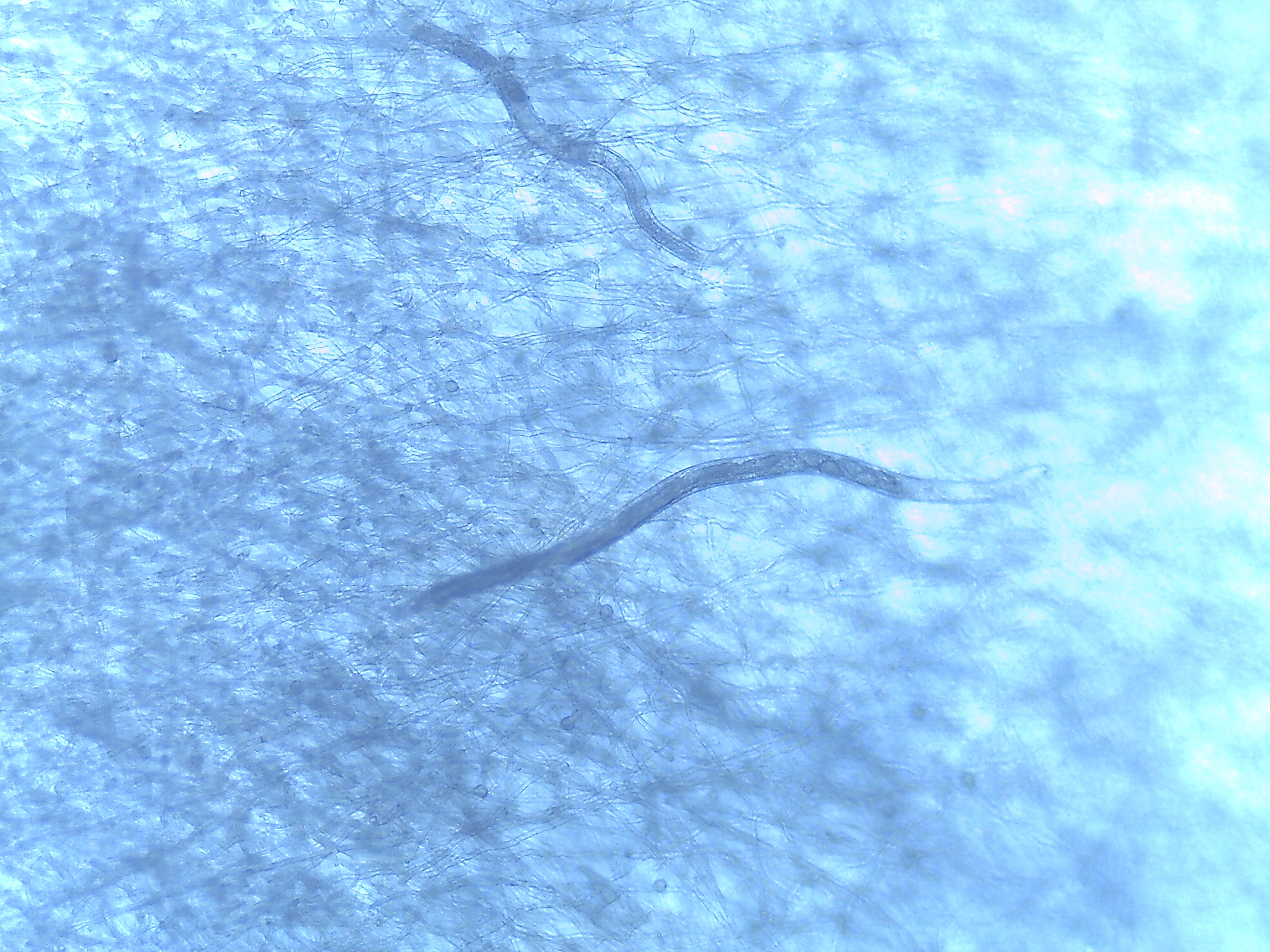
Fungal endobacteria of the genus Mycoavidus show evidence of horizontal transfer between fungal lineages. However, the mechanism of this transmission is unknown. Fungivores in soil, such as Aphelenchus nematodes, may be a vector for these endobacteria. So far, we have detected enrichment of endobacterial DNA relative to fungal nuclear DNA in surface sterilized nematodes, compared to fungal tissue. Future work will include visualizing ingested endobacteria and testing transmission via feeding.
Collaborators: Natalie Golematis, Ashley Barstow
CONSTAXv2: Consensus-based taxonomy assignment for metagenomics
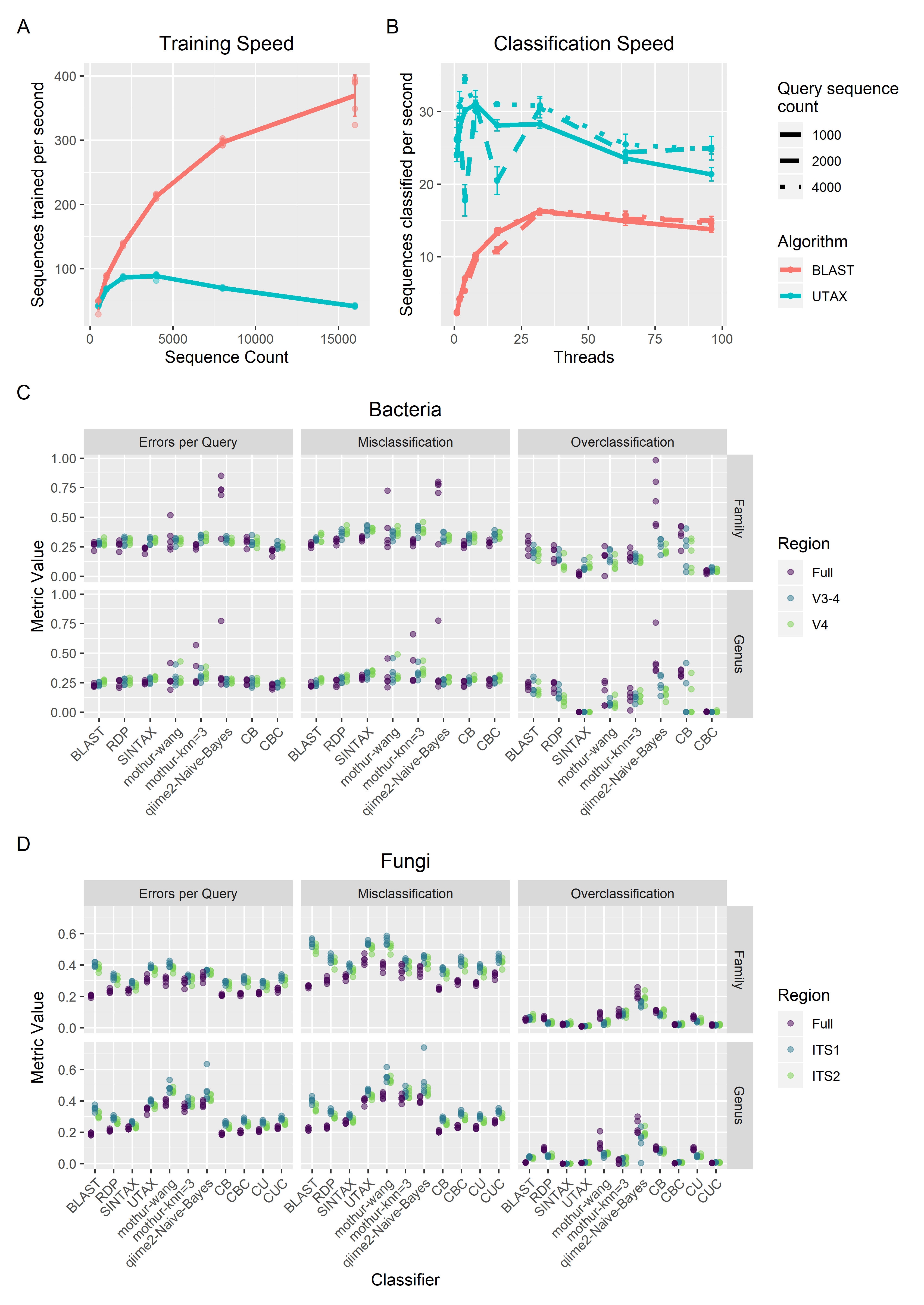
With increasing popularity of metagenomics barcoding studies, researchers require high-throughput taxonomic assignment tools to identify taxa or OTUs from NGS reads. CONSTAXv2 implements a consensus approach, using multiple classifiers (SINTAX, RDP, and UTAX or BLAST) to improve performance. Version 2 implements a new BLAST classification approach, with updated software requirements, an easier-to-use implementation, isolate mapping, and SILVA database compatibility.
Tool Github and Tutorial: https://github.com/liberjul/CONSTAXv2 Preprint: CONSTAX2: Improved taxonomic classification of environmental DNA markers
Collaborators: Dr. Gian Maria Niccolo’ Benucci
Fungal communities of leaf litter
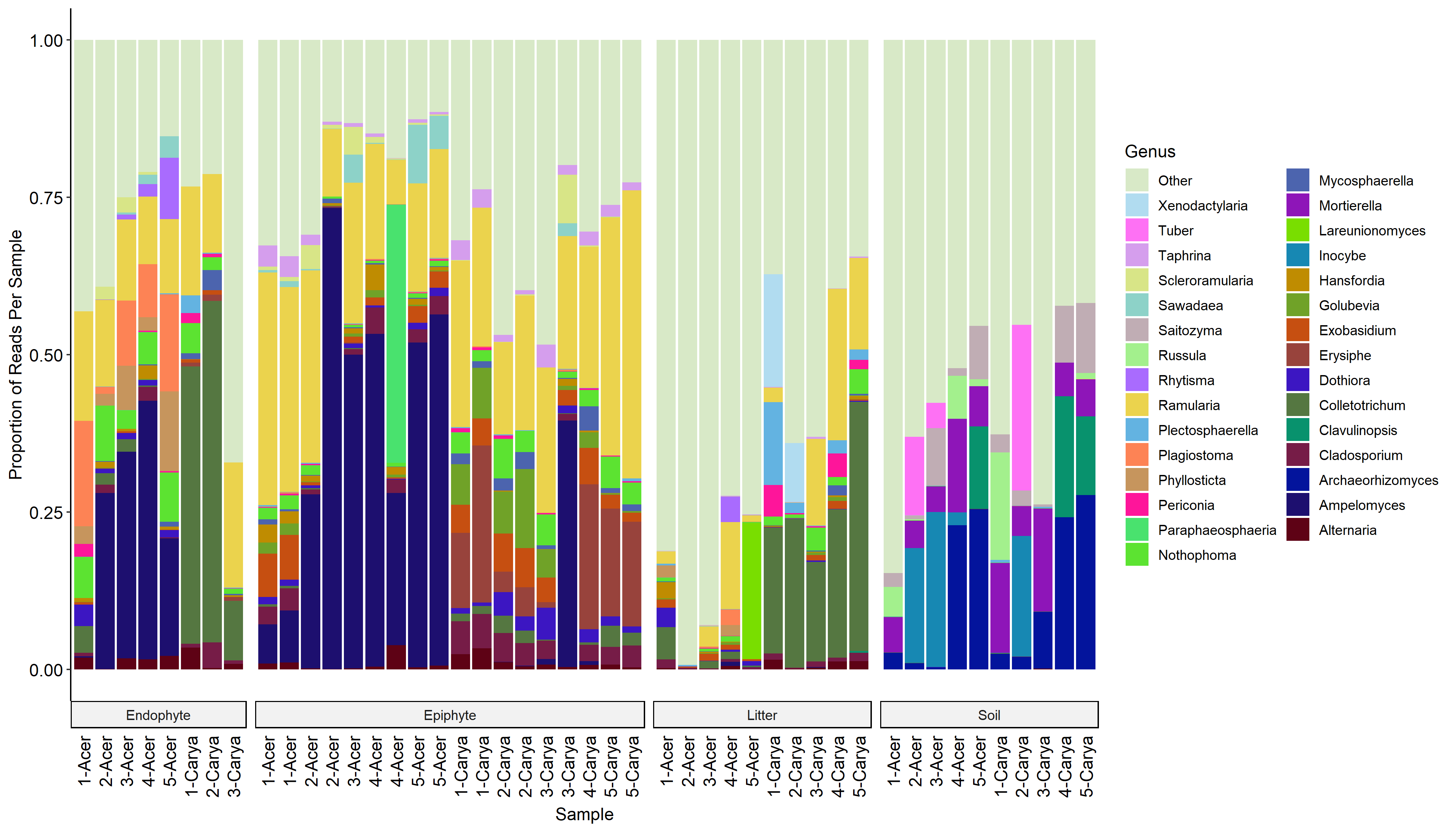 Decomposition of leaf litter in forest ecosystems largely depends on fungi and is a critical process in the terrestrial carbon cycle. Fungi also live in soil and the tree phyllosphere, but the relative contribution of these communities to the leaf litter is unknown. We used ITS amplicon sequencing to characterize and compare fungal communities in leaf litter, soil, leaf surfaces (epiphytes), and leaf internal tissues (endophytes).
Decomposition of leaf litter in forest ecosystems largely depends on fungi and is a critical process in the terrestrial carbon cycle. Fungi also live in soil and the tree phyllosphere, but the relative contribution of these communities to the leaf litter is unknown. We used ITS amplicon sequencing to characterize and compare fungal communities in leaf litter, soil, leaf surfaces (epiphytes), and leaf internal tissues (endophytes).
Collaborators: Jud Van Wyk, Douglas Minier, Anna Stouffer-Hopkins, Reid Longley
Comparative genomics of Mycoplasma-related endobacteria (MRE)
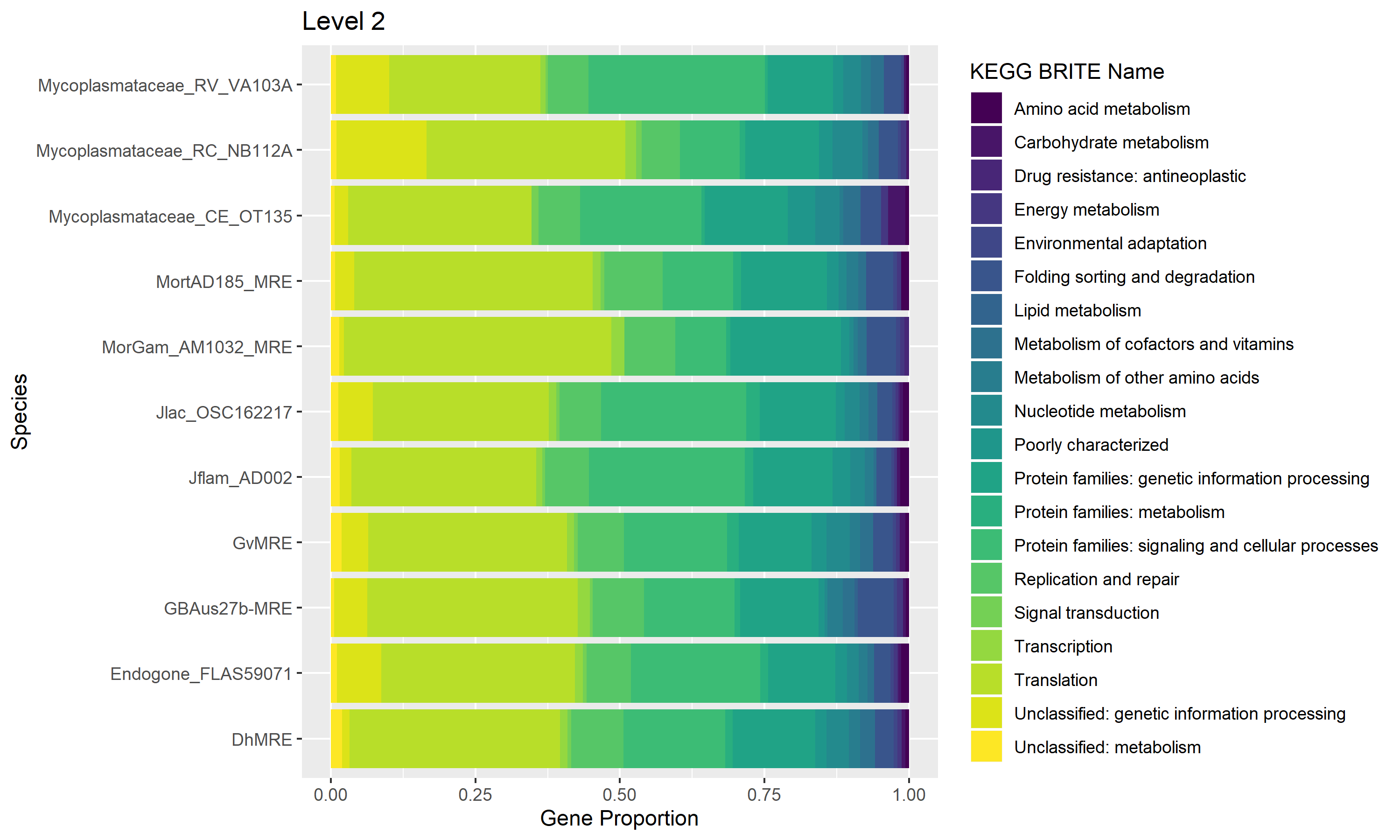
Fungi in the Mucoromycota are known to have Mycoplasma-related bacteria inhabiting their cells. These bacteria are likely obligate endosymbionts, some with highly reduced genomes, and can have impacts on host phenotypes. We are looking for signatures of endosymbiosis in their genomes, and aim to infer the phylogeny of these symbionts and their hosts. While these symbionts may be obligate, there may be horizontal transfer between hosts.
Collaborators: Alicja Okrasińska
Systematics of Mortierellaceae

Mortierellaceae is a family of globally-distributed fungi often associated with soil, insects, or dung. This is a highly diverse group, with undescribed genera and species. We are working to culture new strains from globally-collected substrates, sequence and characterize these strains, and describe new taxa. Some strains in this group may also be useful for arachidonic acid production, microalgae harvesting, or plant symbiosis.
Paper: Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics PDF Collaborators: Dr. Natalie Vande Pol, Natalie Golematis, Audrey Peiker, Abigail Bryson
Automated phenotyping of filamentous fungi
Filamentous fungi are difficult organisms to phenotype, because of the small number of characters which are easily observed. In order to characterize the diversity of groups such as Mortierellaceae, we developed a TensorFlow-based object detection model trained on a manually-curated set of hyphal tip images. This model is accompanied by tools to annotate time-lapse image series infer growth rate by connecting nearest-neighbor annotations between consecutive images.
Tool Github and Tutorial: https://github.com/liberjul/Hyphal_feature_tracking
Collaborators: Natalie Golematis, Audrey Peiker
Algal-fungal co-culture and flocculation
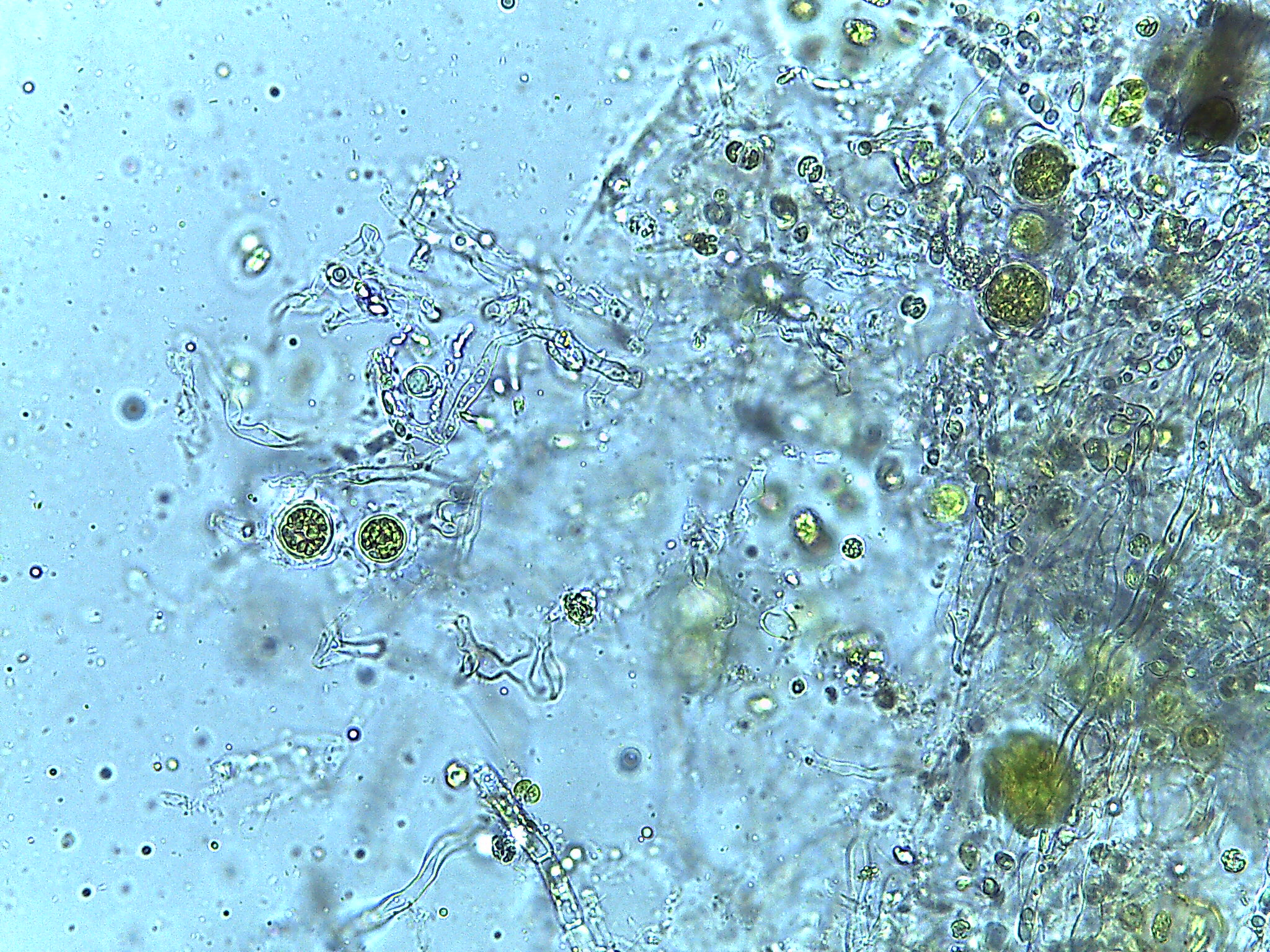
Microalgae are highly productive photosynthetic organisms which have the potential to make valuable products for food, energy, and other industries. Some species of microalgae adhere to Mortierella hyphae which may be exploited for harvesting microalgae. We are working to undercover mechanisms of this interaction.
Review paper: Harvesting microalgae for food and energy products PDF
Collaborators: Dr. Zhi-Yan Du, Conor Bertucci